Overview
The parafac4microbiome package enables R users with an easy way to create Parallel Factor Analysis (PARAFAC) models for longitudinal microbiome data.
-
processDataCube()can be used to process the microbiome count data appropriately for a multi-way data array. -
parafac()allows the user to create a Parallel Factor Analysis model of the multi-way data array. -
assessModelQuality()helps the user select the appropriate number of components by randomly initializing many PARAFAC models and inspecting various metrics of interest. -
assessModelStability()helps the user select the appropriate number of components by bootstrapping or jack-knifing samples and inspecting if the model outcome is similar. -
plotPARAFACmodel()helps visually inspect the PARAFAC model.
This package also comes with three example datasets.
-
Fujita2023: an in-vitro experiment of ocean inocula on peptide medium, sampled every day for 110 days (doi:10.1186/s40168-023-01474-5). -
Shao2019: a large cohort dataset of vaginally and caesarean-section born infants from London (doi:10.1038/s41586-019-1560-1). -
vanderPloeg2024: a small gingivitis intervention dataset with specific response groups (doi:10.1101/2024.03.18.585469).
Documentation
A basic introduction to the package is given in vignette("PARAFAC_introduction") and modelling the example datasets are elaborated in their respective vignettes vignette("Fujita2023_analysis"), vignette("Shao2019_analysis") and vignette("vanderPloeg2024_analysis").
These vignettes and all function documentation can be found on the GitHub pages website here.
Installation
The parafac4microbiome package can be installed from CRAN using:
install.packages("parafac4microbiome")Development version
You can install the development version of parafac4microbiome from GitHub with:
# install.packages("devtools")
devtools::install_github("GRvanderPloeg/parafac4microbiome")Citation
Please use the following citation when using this package:
- van der Ploeg, G. R., Westerhuis, J., Heintz-Buschart, A., & Smilde, A. (2024). parafac4microbiome: Exploratory analysis of longitudinal microbiome data using Parallel Factor Analysis. bioRxiv, 2024-05.
Usage
library(parafac4microbiome)
set.seed(123)
# Process the data cube
processedFujita = processDataCube(Fujita2023,
sparsityThreshold=0.99,
CLR=TRUE,
centerMode=1,
scaleMode=2)
# Make a PARAFAC model
model = parafac(processedFujita$data, nfac=3, nstart=10, output="best", verbose=FALSE)
# Sign flip components to make figure interpretable and comparable to the paper.
# This has no effect on the model or the fit.
model$Fac[[1]][,2] = -1 * model$Fac[[1]][,2] # sign flip mode 1 component 2
model$Fac[[2]][,1] = -1 * model$Fac[[2]][,1] # sign flip mode 2 component 1
model$Fac[[2]][,3] = -1 * model$Fac[[2]][,3] # sign flip mode 2 component 3
model$Fac[[3]] = -1 * model$Fac[[3]] # sign flip all of mode 3
# Plot the PARAFAC model using some metadata
plotPARAFACmodel(model$Fac, processedFujita,
numComponents = 3,
colourCols = c("", "Genus", ""),
legendTitles = c("", "Genus", ""),
xLabels = c("Replicate", "Feature index", "Time point"),
legendColNums = c(0,5,0),
arrangeModes = c(FALSE, TRUE, FALSE),
continuousModes = c(FALSE,FALSE,TRUE),
overallTitle = "Fujita PARAFAC model")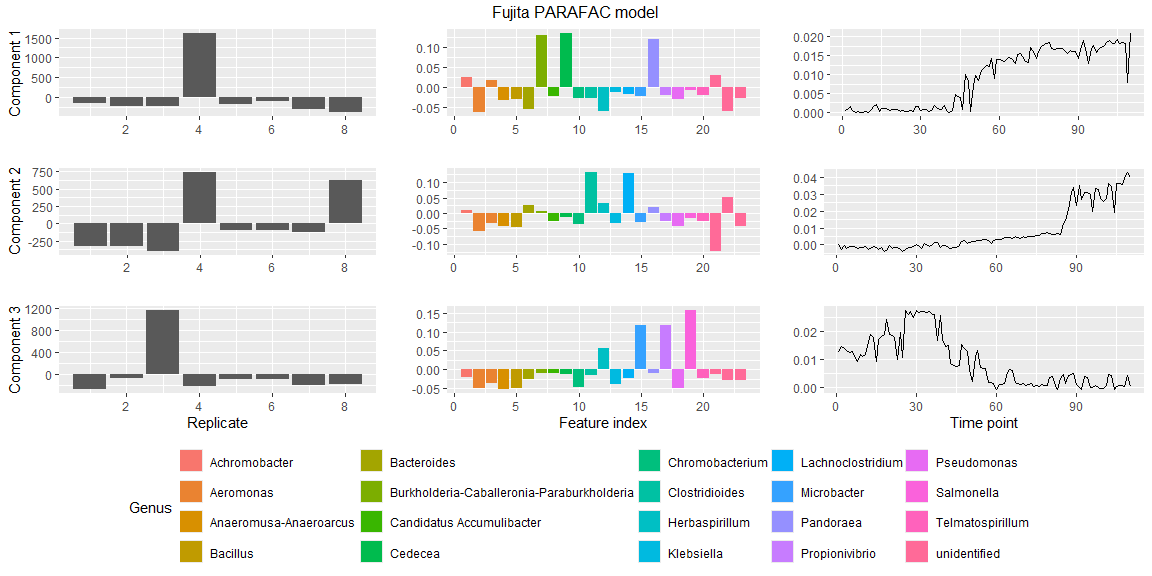
Getting help
If you encounter an unexpected error or a clear bug, please file an issue with a minimal reproducible example here on Github. For questions or other types of feedback, feel free to send an email.
